Abstract
Hematopoiesis involves orchestrated differentiation of hematopoietic stem cells (HSCs) and lineage-restricted progenitors to mature blood cells. Different populations in this hierarchy vary in function, size, proliferation, and protein synthesis rates. It is also known that different normal hematopoietic cell types differ in the rates of ribosomal RNA (rRNA) transcription, the key rate-limiting step in ribosome biogenesis that occurs in the nucleolus. Leukemic blast cells have long been known to have enlarged nucleoli, indicating aberrantly increased rRNA transcription and ribosome biogenesis. Ribosome biogenesis is an energy-intensive process, beginning with the transcription of multi-copy rDNA genes by RNA polymerase I (Pol I) to produce 47S precursor rRNA (pre-rRNA), which is further processed into mature 18S, 5.8S, and 28S rRNA and assembled with 5S rRNA and 80 different ribosomal proteins to form mature ribosomes (Fig A). Because rRNA transcription is often considered a housekeeping process, the variations in its cell-type and disease-specific rates have not been studied, leading to a lack of understanding of the mechanisms underlying its regulation.
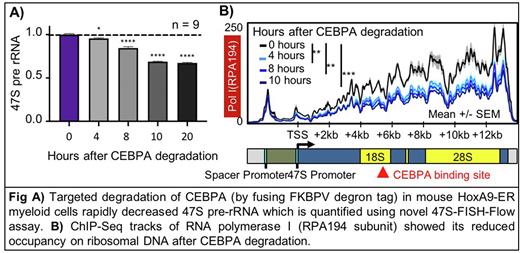
To identify cell-type-specific regulators of rRNA transcription in hematopoiesis, we compiled a list of 249 hematopoietic transcription factors (TFs) and epigenetic factors, and mapped 2200 publicly available ChIP-Seq datasets for these factors to rDNA, creating the first atlas of hematopoietic TF-rDNA binding. Through this atlas, we identified that CEBPA shows consistent and abundant binding to human and mouse rDNA at a conserved, previously unknown motif (Fig B). CEBPA is an essential myeloid lineage-specific TF whose knockout leads to complete loss of granulocyte-monocyte progenitors (GMPs) and all downstream myeloid lineage cells. CEBPA also shows hotspot mutations in 10% of AML patients. To characterize the role of CEBPA in rRNA transcription, we used a mouse HoxA9-ER cell line (which closely resembles GMPs), and biallelically fused FKBP degron into endogenous loci of the Cebpa gene, enabling rapid degradation of CEBPA protein using small molecule ligand dTAGV-1 (Fig C, D). To quantify effects on rRNA transcription, we developed a novel assay named '47S-FISH-Flow’ that involves flowcytometric quantification of fluorescent probes hybridized to nascent rRNA in the nucleolus (Fig E, F). We found that depleting CEBPA caused a rapid reduction in 47S rRNA level, plateauing at 35% reduction within hours (Fig G). Decrease in rRNA production was followed by reduction in nucleolar size, reduction in mature ribosome subunit abundance, and reduced growth (Fig H, I). To understand the molecular function of CEBPA on rDNA machinery, we performed time course ChIP-Seq on key rDNA transcriptional machinery, and found that CEBPA degradation specifically reduced occupancy of Pol I along with its initiation factor RRN3 on rDNA (Fig J).
In summary, we have found a novel role for the extensively studied myeloid lineage-specific TF CEBPA in directly binding rDNA and regulating rRNA transcription. Our results, and the tools and experimental systems we have developed, shed light on an important and largely unexplored aspect of hematopoietic biology: the regulation of rRNA transcription by lineage-specific hematopoietic TFs.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal